Research Project
NISS joined the Clinical Proteomic Technologies for Cancer research initiative that focuses on technologies for cancer biomarker discovery. The overall goal was to foster the building of an integrated foundation of proteomic technologies, data, reagents and reference materials, and analysis systems. This will systematically advance the application of protein science to accelerate discovery and research in cancer. Progress in cancer proteomic research has continually been hampered by a lack of standardized technologies and methodologies which are critically needed for effective discovery and validation of proteins and peptides as biomarkers. The challenges to the Clinical Proteomic Technology Assessment for Cancer (CPTAC),a collaborative network of proteomic centers plus the National Institute of Standards and Technology, are to (1) conduct a rigorous assessment of two major technologies for protein and peptide analysis – mass spectrometry (both shotgun and MRM) and affinity capture platforms; and (2) develop and validate technical tools for general use in proteomics that ensure reliable and reproducible results, analyses and interpretations.
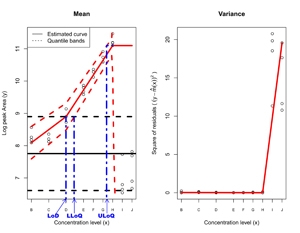
Statistical designs were constructed using algebra theory to achieve sparse high dimensional balance of relevant factors for multidimensional responses; designs were translated into operating instructions for preparation of blinded samples. Functional analysis was used with non-parametric smoothing under shape constraints to develop calibration methodology with optimality properties for mass spectrometry data. Also a second new, less computationally intensive, calibration algorithm was developed for rapid, highdimensional calculations under the common special case of linear (with or without log transformation) response over an instrument’s dynamic range. High dimensional data analytic techniques were applied to identify and quantitate sources of variation in mass spectrometry for proteomics. New statistically-based diagnostics were developed for monitoring and analyzing these proteomic data.
All new methodologies together with best available practices in design and in analysis were applied to multiple experiments, from small investigations of specific stages of the system (e.g., digestion) to large scale, complex studies (e.g., >120 peptides, 4 transitions, 9 – 16 concentrations, 4 – 17 laboratories, 3-7 platforms).
CPTAC Institutions
Broad Institute of MIT and Harvard
Memorial Sloan-Kettering Cancer Center
Purdue
UC-San Francisco/Lawrence Berkeley National Laboratory and
Buck Institute
Vanderbilt School of Medicine
Provide state of the art statistical methodology, statistical analysis and process evaluation for CPTAC experimental proteomics data; Design complex multi-institutional, multivariate experiments and calibrations for the CPTAC network; Develop statistical methodologies to advance proteomic science.
NISS senior Directors, experienced in biostatistics, “omics” computational technology, metrology and calibration: Nell Sedransk and Stan Young. NISS postdoctoral fellows with specific expertise in bioinformatics, statistical computation, and statistical applications for “omics” and other large scale data: Xingdong Feng, Jessie Xia, and Xia Wang.
